Haplotype association studies
© NeuroSpin/CEA.
Grey matter thickness is known to shrink with aging in both diseased and normal brains. A related effect is the cortical sulcus widening : the width of a sulcus can be estimated using a feature called opening. Sulcus opening is shown to be robustly related to grey matter thickness and does not require spatial normalization nor regional atlas.
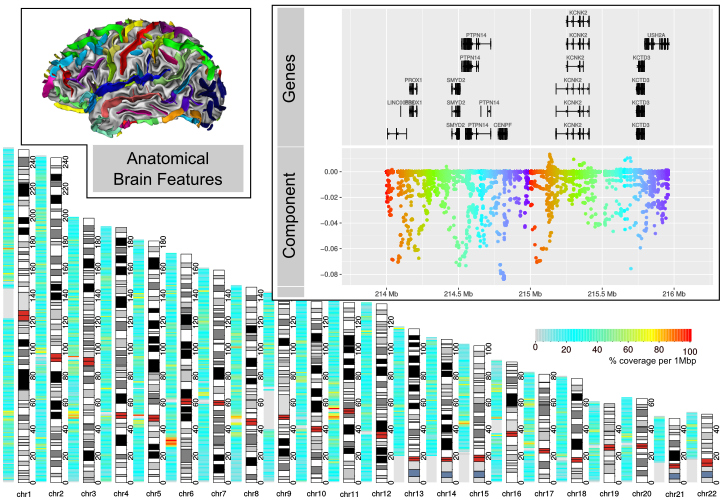
Imaging genetics studies of large control cohorts such as UK Biobank enable to assess the range of normal variations in brain structures. Heritability studies pointed to a dozen sulci that appeared to be under strong genetic control. Using GWAS, our lab has identified a reproducible genetic marker associated with the opening of the left, posterior, Calloso-Marginal sulcus: the width of several cortical sulci is associated with a variant in the upstream region of KCNK2 gene even if this effect is corrected with age.
However, GWAS use a univariate approach and as such, suffer from several drawbacks, in particular the use of an unduly conservative multiple test correction and the fact that the correlation structure of the genome is not accounted for. In the context of complex traits, where individual variant effect size is expected to be small, only SNPs that are frequent in the population can significantly be associated with a phenotype under study. Moreover, univariate analyses are unable to model or predict the role of a genetic variant within the genomic region. Finally, univariate approaches are inadequate in situations where a set of variants are jointly associated to multiple phenotypes (pleiotropy).
Using a multivariate approach, we propose to alleviate these drawbacks by simultaneously analyzing hundred related phenotypes and modeling interactions between genetic variants within the same genomic segment. The genetic variants we consider are sets of SNPs of known phase called haplotypes, taken from the upstream region of KCNK2 gene. So far, multivariate analysis in imaging genetics have never used haplotypes. Our method was able to recover the expected association signal and uncover new associations between imaging data and genetic variants.
Karkar, S., Gloaguen, A., Le Guen, Y., Pierre-Jean, M., Dandine-Roulland, C., Le Floch, E., … Frouin, V. (2019). Multivariate haplotype analysis of 96 sulci opening for 15,612 UK-Biobank subjects. In ISBI 2019 - Proceedings of the IEEE International Symposium on Biomedical Imaging. Venice, Italy. Retrieved from https://hal-cea.archives-ouvertes.fr/cea-02016827